Pre-Processing Steps
I wanted to talk about how we pre-process data in the lab, and some other approaches
HCP Approach
The information here is taken from the reference manual for the last release of HCP data titled HCP_S1200 (here)
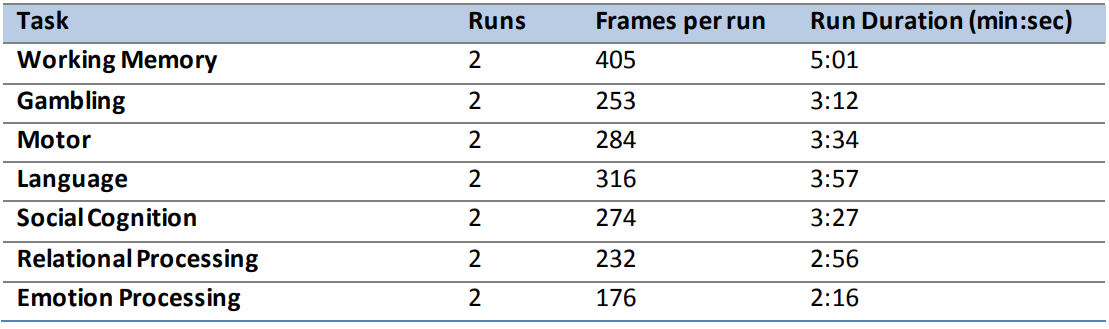
Some notable points are that that the resting state totals 57 minutes!

The other tasks that are included are below:
THE reference for the HCP pre-processing pipline is the below publication:
The paper notes that most researchers should probably work with the pre-proccessed datasets to not have to deal with esoteric issues such as the “correction of gradient nonlinearity distortion in images acquired with oblique slices”. Overall, there are 6 goals of the pipeline:
- Remove spatial artifacts and distortions
- Generate cortical surfaces, sgmentations, and myelin maps
- Make the data easily viewable in the workbench software
- Generate precise within-subject cross-modal registrations
- Handle surface and volume cross-subject registrations to standard volume and surface spaces
- Make the data available in CIFTI format
These major goals are accomplished through 6 minimal preprocessing pipelines.
Overview
The pipelines can be thought of as dealing with structural and functional data. We will first deal with structural.
PreFreeSurfer
The first structural pipeline PreFreeSurfer produces an undistorted “native” structural volume space for each subject, aligns the T1w and T2w images, performs a bias field correction, registers the subjects native structural volume to MNI space. The outputs of the PreFreeSurfer pipeline are organized into folders called T1w that contains native volume space images and a second folder called MNINonLinear that contains the MNI space images.
FreeSurfer
This can be thought of as a fine-tuned version of FreeSurfer’s recon-all pipeline. The main changes are that the structural volumes are downsampled to 1mm isotropic voxels, and the brain mask from the previous step is used as an input. The final steps of this algorithm are pial surface generation and morphometric measurements.
PostFreeSurfer
This takes the proprietary output formats of FreeSurfer and converts them to stndard NIFT and GIFTI formats. Of note, one of these files (wmparc) is dilated and eroded to produce an accuracte subject specific brain mask of the gray and white matter. This pipeline also registers the individual subjects native-mesh surfaces to the Conte69 population average surface (32k per hemisphere)
This is then followed by the PostFreeSurfer pipeline which produces all of the NIFTI volume and GIFTI surface files necessary for viewing the data in Connectome Workbench
The first functional pipeline, fMRIVolume, removes spatial distortions, realigns volumes, registeres the fMRI data to structural, reduces bias field, normalizes the 4D image to a global mean, and makss the data with the final brain mask. Note, this pipeline does no overt smoothing. The second functional pipeline, fMRISurface brings the timeseries from the volume into the CIFTI grayordinate standard space.
In the HCP pipeline, high resolution T1w and T2w images are acquired in order to create accurate cortical surface and myelin maps. Any repeated runs of T1w and T2w images are aligned with a 6 DOF rigid body transformation using FSL’s FLIRT. The average is then rigidly transformed to MNI space and alignes the AC, the AC-PC line and the inter-hemispheric plane (but maintains original size and shape of the brain).
To understand the preproccessing pipeline, we have to go to the WashU github page (here).
Waiting to sync
fMRIVolume
This requires completion of the HCP structural pipelines. There are two types of distortion corrections: Gradient and EPI distortion corrections.
fMRISurface
The goal of this pipeline is to take a volume timeseries and map it to the standard CIFTI grayordinate spacce.
Taking from the paper:
The end results are 2 mm FWHM smoothed surface timeseries on a standard mesh in which the vertex numbers correspond to spatially matched locations across subjects